Friday, December 29, 2006
More fabrication techniques
In the late 1990s, Tom Knight at MIT worked on something he called microbial engineering, the intentional redesign of simple (prokaryotic) bacteria, which has resulted in MIT's Biological Parts Project. The idea is to identify re-usable components that can be included in rationally designed microorganisms to perform various functions.
This idea is not without precedent: in 1978, Genentech re-engineered E. coli bacteria to produce inexpensive human insulin, vital to the survival of diabetes patients. Previously insulin had been extracted from ground-up organs of farm animals at considerably greater expense. The 1978 work did not have access to a catalog of biological parts or many of the techniques and other knowledge infrastructure that will grow up around the MIT work.
In an earlier posting I described some very interesting work being done by Christian Schafmeister, who is assembling monomer chains to create structures with specific, controllable, and reasonably rigid shapes. He is developing a collection of 15 or 20 monomers, and perhaps that number will grow over time, which can be strung together using synthetic chemistry techniques. Schafmeister has an article in this month's Scientific American.
DNA origami exploits the very selective self-stickiness of DNA. It is likely that DNA (which can be created in any desired sequence) will become a very flexible framework on which to position molecules. Proteins can also be engineered, provided we can predict how they will fold, and this should be a solvable problem if we restrict ourselves to a subset of well-understood proteins. Many proteins like to cling to DNA at very specific locations. A combined approach using a DNA scaffolding, with attached proteins to provide local functionality, could yield very interesting results.
This idea is not without precedent: in 1978, Genentech re-engineered E. coli bacteria to produce inexpensive human insulin, vital to the survival of diabetes patients. Previously insulin had been extracted from ground-up organs of farm animals at considerably greater expense. The 1978 work did not have access to a catalog of biological parts or many of the techniques and other knowledge infrastructure that will grow up around the MIT work.
In an earlier posting I described some very interesting work being done by Christian Schafmeister, who is assembling monomer chains to create structures with specific, controllable, and reasonably rigid shapes. He is developing a collection of 15 or 20 monomers, and perhaps that number will grow over time, which can be strung together using synthetic chemistry techniques. Schafmeister has an article in this month's Scientific American.
DNA origami exploits the very selective self-stickiness of DNA. It is likely that DNA (which can be created in any desired sequence) will become a very flexible framework on which to position molecules. Proteins can also be engineered, provided we can predict how they will fold, and this should be a solvable problem if we restrict ourselves to a subset of well-understood proteins. Many proteins like to cling to DNA at very specific locations. A combined approach using a DNA scaffolding, with attached proteins to provide local functionality, could yield very interesting results.
Thursday, December 07, 2006
Molecular dynamics simulation of small bearing design
This video was created using the simulation facilities of NanoEngineer-1 (see http://www.nanoengineer-1.com), together with open-source animation tools like Pov-RAY, ImageMagick, and mpeg2encode. This is a simulation of the molecular bearing design on page 298 of "Nanosystems" by Eric Drexler. When viewed at 0.15 picoseconds per second of animation, thermal motion of atoms (particularly hydrogens) is visible. At 0.6 picoseconds per second, thermally excited mechanical resonances of the entire structure are seen. At 6 picoseconds per second, the rotation of the shaft (one rotation every 200 psecs) becomes apparent.
Update: On more careful analysis we discovered that the temperature is incorrectly represented in this video. The atoms should shake more violently to represent an ambient temperature of 300 Kelvin (ordinary room temperature). The vibrations you see in the video correspond to about 70 Kelving (very chilly). In spite of the more violent thermal vibrations, the structure remains chemically stable and mechanically workable at room temperature.
Friday, October 27, 2006
Nature article on molecular motors
A Nature article on molecular motors found in biology. I'm not sure of the date, I found this on the Advanced Nanotechnology Blog maintained by Brian Wang.
Thursday, August 03, 2006
Automated design
Design is a search problem. A product or machine has some kind of specification or instructions, and you want to find the best possible specification to suit some purpose. The space of all possible specifications is usually too large to be searched exhaustively. The usual response to this is to exercise human cleverness - what the inventor in the garage does. An alternative is to search the design space using computer algorithms.Why would you do that? Isn't it fun to invent stuff? It is, but the stuff we invent these days is getting so complicated that sometimes human cleverness might not suffice to solve a design challenge. Nanotech stuff will be orders of magnitude more complicated than anything we can make today. So it's good to take a look at this approach.
Automated design is the application of global optimization to design problems using techniques like simulated annealing, genetic algorithms, and ant colony optimization to generate candidate problem solutions, and computer simulations to evaluate the fitness of the candidates. Genetic algorithms are the most widely known of these techniques. Here is a discussion of GAs by John Holland, one of the early pioneers in the field.
There has been a lot of research and development applying GAs to
engines and other machinery. Here are two applications of GAs to optimize the design of diesel engines. Somebody else optimized a valvetrain., and somebody else, an exhaust manifold. There is also work on a flywheel made of composite materials, and some work on reducing engine emissions by optimizing the chemical reaction rate of the fuel. In a more nanotech-ish vein, there is some work on the molecular design of novel fibers and polymers.
Here are two applications of GAs to optimize the design of diesel engines. Somebody else optimized a valvetrain., and somebody else, an exhaust manifold. There is also work on a flywheel made of composite materials, and some work on reducing engine emissions by optimizing the chemical reaction rate of the fuel. In a more nanotech-ish vein, there is some work on the molecular design of novel fibers and polymers.
 One of the more interesting efforts in this field is John Koza's invention machine, which applies GAs to a very wide variety of design problems, and which has produced a number of new patents for designs that did not originate in human imaginations.
One of the more interesting efforts in this field is John Koza's invention machine, which applies GAs to a very wide variety of design problems, and which has produced a number of new patents for designs that did not originate in human imaginations.

NASA used genetic algorithms to design microwave antennas for the ST5 mission to measure the Earth's magnetosphere. There is some discussion of this work on Wikipedia.
While genetic algorithms are the most widely known of this class of algorithms, simulated annealing has a decades-long history of success in the placing and routing of FPGAs and custom integrated circuits. Other applications for global optimization include scheduling and resource allocation.
Random links
Automated design is the application of global optimization to design problems using techniques like simulated annealing, genetic algorithms, and ant colony optimization to generate candidate problem solutions, and computer simulations to evaluate the fitness of the candidates. Genetic algorithms are the most widely known of these techniques. Here is a discussion of GAs by John Holland, one of the early pioneers in the field.
There has been a lot of research and development applying GAs to
engines and other machinery.
 Here are two applications of GAs to optimize the design of diesel engines. Somebody else optimized a valvetrain., and somebody else, an exhaust manifold. There is also work on a flywheel made of composite materials, and some work on reducing engine emissions by optimizing the chemical reaction rate of the fuel. In a more nanotech-ish vein, there is some work on the molecular design of novel fibers and polymers.
Here are two applications of GAs to optimize the design of diesel engines. Somebody else optimized a valvetrain., and somebody else, an exhaust manifold. There is also work on a flywheel made of composite materials, and some work on reducing engine emissions by optimizing the chemical reaction rate of the fuel. In a more nanotech-ish vein, there is some work on the molecular design of novel fibers and polymers. One of the more interesting efforts in this field is John Koza's invention machine, which applies GAs to a very wide variety of design problems, and which has produced a number of new patents for designs that did not originate in human imaginations.
One of the more interesting efforts in this field is John Koza's invention machine, which applies GAs to a very wide variety of design problems, and which has produced a number of new patents for designs that did not originate in human imaginations.
NASA used genetic algorithms to design microwave antennas for the ST5 mission to measure the Earth's magnetosphere. There is some discussion of this work on Wikipedia.
While genetic algorithms are the most widely known of this class of algorithms, simulated annealing has a decades-long history of success in the placing and routing of FPGAs and custom integrated circuits. Other applications for global optimization include scheduling and resource allocation.
Random links
Monday, July 24, 2006
Spimes
Bruce Sterling gave an interesting talk at a SIGGRAPH conference in 2004. He described two kinds of human artifacts, blobjects and spimes. Blobjects are simply artifacts that have been designed with modern CAD systems, so their shapes are more curvy and sexy than the same-functioned artifacts of past generations. Examples are the iMac and the new VW beetle.
The spime is a different beast. It is jam-packed full of information technology. It has RFID or Bluetooth to talk to nearby computers (or maybe other spimes). It has GPS so it knows where on Earth it is. It knows how to connect to the Internet. It willingly participates in data mining efforts by Google and other search engines and advertisers. In addition to being designed with a CAD system, it might be manufactured with rapid prototyping techniques such as 3D printers.
Sterling's predictions about the spimes' use of information are cynical. They are programmed by the corporations that built them. They collect consumer demographics information about the people who buy and use them. Their first allegiance is to their manufacturer. They are smart enough that the distinction has teeth - the hand drill I bought at Sears does not change its behavior to act in Sears' best interests rather than mine.
If spimes aren't nanotechnology, why am I writing about them in a nanotech blog? Because they shake loose my thinking about what products could be. I hadn't thought about ANY of this stuff before I read the transcript of Sterling's talk. My cell phone today has way more computing power than the Apollo guidance computer had. When a ballpoint pen has way more computing power than my cell phone has today, of course somebody will program it to do things like this.
The spime is a different beast. It is jam-packed full of information technology. It has RFID or Bluetooth to talk to nearby computers (or maybe other spimes). It has GPS so it knows where on Earth it is. It knows how to connect to the Internet. It willingly participates in data mining efforts by Google and other search engines and advertisers. In addition to being designed with a CAD system, it might be manufactured with rapid prototyping techniques such as 3D printers.
Sterling's predictions about the spimes' use of information are cynical. They are programmed by the corporations that built them. They collect consumer demographics information about the people who buy and use them. Their first allegiance is to their manufacturer. They are smart enough that the distinction has teeth - the hand drill I bought at Sears does not change its behavior to act in Sears' best interests rather than mine.
If spimes aren't nanotechnology, why am I writing about them in a nanotech blog? Because they shake loose my thinking about what products could be. I hadn't thought about ANY of this stuff before I read the transcript of Sterling's talk. My cell phone today has way more computing power than the Apollo guidance computer had. When a ballpoint pen has way more computing power than my cell phone has today, of course somebody will program it to do things like this.
Thursday, April 27, 2006
Creepy crawlies 2
The gray goo scenario is not an immediate threat. Evolution of nanomachines can be prevented, and gray goo replicators can simply not be designed, as long as everybody agrees to those rules. If that were that, we could prevent the gray goo scenario forever. But not everybody will agree. Some, having loudly agreed in public, will quietly break the rules in private. But real live gray goo won't become possible very soon.
There are lots of "interesting" lesser threats.
Another possibility is to educate ourselves about the possible range of weapons, in the hopes of designing effective defenses. This is a much more technically tractable problem, and it's one of the reasons I work for Nanorex. Our software can help people to explore the space of possible threats and defenses more quickly, and to create an active research literature.
Is an active literature a good idea? Would it not be an enabler for those who wish to do harm? Should it not be suppressed or at least discouraged?
This is like the relinquishment question. If the bad guys have an active research literature and the good guys don't, then the first bad guy attack could reduce the good guys to a state where they could never again hope to meaningfully protect themselves.
Ideally, thoroughly effective defenses would be deployed everywhere, long before the first offensive weapons appear. I hope it goes that way. If we're not so lucky, there may turn out to be such a diversity of possible offensive weapons that "effective defenses" aren't practical.
There are lots of "interesting" lesser threats.
At the time of the Pontiac rebellion in 1763, Sir Jeffrey Amherst, the Commander-in-Chief of the British forces in North America, wrote to Colonel Henry Bouquet: 'Could it not be contrived to send smallpox among these disaffected tribes of Indians? We must use every stratagem in our power to reduce them.' The colonel replied: 'I will try to inoculate the [Native American tribe] with some blankets that may fall in their hands, and take care not to get the disease myself.' Smallpox decimated the Native Americans, who had never been exposed to the disease before and had no immunity.What we can expect in the near term is biological warfare, evolving along lines similar to biological warfare today, and with similar motives. If we are interested in a future world that is benign, we can try to remove the incentives for biological warfare, so that there is as little of it as possible. This is an area where I myself can offer no particular insight. There have been many many different manifestations of human evil in recent decades and centuries. I would like to think I could entrust politicians and diplomats to go about its prevention, but they are usually the ones who start the next one. This is a thorny problem involving culture clashes, economics, religion, and many other topics beyond the scope of this blog.Silent Weapon: Smallpox and Biological Warfare by Colette Flight writing for bbc.co.uk/history
Another possibility is to educate ourselves about the possible range of weapons, in the hopes of designing effective defenses. This is a much more technically tractable problem, and it's one of the reasons I work for Nanorex. Our software can help people to explore the space of possible threats and defenses more quickly, and to create an active research literature.
Is an active literature a good idea? Would it not be an enabler for those who wish to do harm? Should it not be suppressed or at least discouraged?
This is like the relinquishment question. If the bad guys have an active research literature and the good guys don't, then the first bad guy attack could reduce the good guys to a state where they could never again hope to meaningfully protect themselves.
Ideally, thoroughly effective defenses would be deployed everywhere, long before the first offensive weapons appear. I hope it goes that way. If we're not so lucky, there may turn out to be such a diversity of possible offensive weapons that "effective defenses" aren't practical.
Wednesday, April 26, 2006
Lessons from software development
I've been a software engineer for ten years now. Software engineers build and maintain very complex systems, so complex that no single engineer can grasp the whole thing in all its details all at once. Coming from electrical engineering, I found that a humbling new experience.
Software engineering looks a particular way, because bits are much much cheaper than transistors. Software products are generally much more complex than hardware products. So software engineering has a richer set of tools for managing complexity than hardware engineering has.
Testing is hugely important. Engineers test products, and the more testing they do earlier, the fewer bugs they need to fix later. If you've got a system that's incomprehensibly complex, you can still understand how to test it.
Modularity is an example of information hiding. Designs are made up of individually comprehensible pieces or modules. Each piece has complexity inside and simplicity outside. The simple outside part, the interface, determines how that piece works with its neighbors.
Software engineering looks a particular way, because bits are much much cheaper than transistors. Software products are generally much more complex than hardware products. So software engineering has a richer set of tools for managing complexity than hardware engineering has.
Testing is hugely important. Engineers test products, and the more testing they do earlier, the fewer bugs they need to fix later. If you've got a system that's incomprehensibly complex, you can still understand how to test it.
Modularity is an example of information hiding. Designs are made up of individually comprehensible pieces or modules. Each piece has complexity inside and simplicity outside. The simple outside part, the interface, determines how that piece works with its neighbors.
Tuesday, April 25, 2006
nanoENGINEER-1, alpha 7 release
The Nanorex team is proud to announce the release of nanoENGINEER-1 Alpha 7, the seventh major release of nanoENGINEER-1. This is the first major release since Alpha 6 was announced on August 17, 2005. Alpha 7 has many improvements and new features.
Undo/Redo
Alpha 7 now includes a new Undo/Redo system making it easy to undo accidental changes to models.
Simulator/Minimizer Improvements
The nanoENGINEER-1 simulator and minimizer have undergone major revisions since Alpha 6. A7 introduces our new and improved molecular force field. We have expanded the list of parameters from our initial testing set to include more atoms and bond types from among the main group elements. It is our intent to have all possible bond types for the first three rows of the main group of the periodic table (B-F, Al-Cl, Ga-Br, and H with all chemically possible combinations of single, double, and triple bonding) accounted for with the completion of the simulator work, except for bond torsion terms, which are planned for Alpha 8. Once this initial set is complete, we will welcome requests for other bond types.
To ensure the accuracy of the minimizer and, consequently, our new molecular force field model, we are developing tests which compare the minimizer output to quantum mechanical results performed at the same high level of theory (B3LYP/6-31+G(d,p)) as the force field itself. These tests have been run on a variety of small structures to cover as many of the force field parameters as possible.
Improved Modeling Interface
Making nanoENGINEER-1 easy to learn and use is one of our highest priorities. With Alpha 7, building models has never been easier and more intuitive. Chemists using commercial molecular modeling programs such as Chem3D, Spartan or Accelrys will find nanoENGINEER-1 a breeze and enjoy its ability to manipulate large chemical species just as easily as small molecules. Mechanical engineers experienced with traditional CAD programs like SolidWorks or Pro/ENGINEER will feel right at home creating and viewing models even though their knowledge of chemistry may be limited.
Wiki-based Help
A new Wiki web site for nanoENGINEER-1 hosts a set of on-line Help pages that can be accessed by pressing the F1 Key while the cursor is hovering over objects in the nanoENGINEER-1 user interface. Users are able (and encouraged) to create a Wiki user account and add or edit the Help pages themselves. We anticipate that over time the Wiki will become an invaluable resource for users of nanoENGINEER-1.
Improved Graphics
Graphics quality and POV-Ray support has been improved significantly. We’ve included animation when switching between views, specular highlighting and improvements to the general graphics and lighting control system that give nanoENGINEER-1 a professional luster it lacked in previous versions.
Nano-Hive Support
nanoENGINEER-1 can serve as a general purpose molecular modeling front-end to Nano-Hive, a nanospace simulator written by Brian Helfrich. Working together, nanoENGINEER-1 allows users to interactively define, calculate and visualize 2D electrostatic potential (ESP) images of molecular models. To see an example of this new capability, check out the ESP Image jig in the nanoENGINEER-1 Gallery.

Mac OS X
Alpha 7 does not yet work on Intel-based Mac systems. We are working on this and hope to support Intel-based Mac systems in the coming weeks. We also just discovered a problem plotting nanoENGINEER-1 simulation trace files using GNUplot on Mac OS X. We will be fixing this problem in the next day or two and posting a new MacOS install package in the Download Area.
Undo/Redo
Alpha 7 now includes a new Undo/Redo system making it easy to undo accidental changes to models.Simulator/Minimizer Improvements
The nanoENGINEER-1 simulator and minimizer have undergone major revisions since Alpha 6. A7 introduces our new and improved molecular force field. We have expanded the list of parameters from our initial testing set to include more atoms and bond types from among the main group elements. It is our intent to have all possible bond types for the first three rows of the main group of the periodic table (B-F, Al-Cl, Ga-Br, and H with all chemically possible combinations of single, double, and triple bonding) accounted for with the completion of the simulator work, except for bond torsion terms, which are planned for Alpha 8. Once this initial set is complete, we will welcome requests for other bond types.To ensure the accuracy of the minimizer and, consequently, our new molecular force field model, we are developing tests which compare the minimizer output to quantum mechanical results performed at the same high level of theory (B3LYP/6-31+G(d,p)) as the force field itself. These tests have been run on a variety of small structures to cover as many of the force field parameters as possible.
Improved Modeling Interface
Making nanoENGINEER-1 easy to learn and use is one of our highest priorities. With Alpha 7, building models has never been easier and more intuitive. Chemists using commercial molecular modeling programs such as Chem3D, Spartan or Accelrys will find nanoENGINEER-1 a breeze and enjoy its ability to manipulate large chemical species just as easily as small molecules. Mechanical engineers experienced with traditional CAD programs like SolidWorks or Pro/ENGINEER will feel right at home creating and viewing models even though their knowledge of chemistry may be limited.Wiki-based Help
A new Wiki web site for nanoENGINEER-1 hosts a set of on-line Help pages that can be accessed by pressing the F1 Key while the cursor is hovering over objects in the nanoENGINEER-1 user interface. Users are able (and encouraged) to create a Wiki user account and add or edit the Help pages themselves. We anticipate that over time the Wiki will become an invaluable resource for users of nanoENGINEER-1.Improved Graphics
Graphics quality and POV-Ray support has been improved significantly. We’ve included animation when switching between views, specular highlighting and improvements to the general graphics and lighting control system that give nanoENGINEER-1 a professional luster it lacked in previous versions.Nano-Hive Support
nanoENGINEER-1 can serve as a general purpose molecular modeling front-end to Nano-Hive, a nanospace simulator written by Brian Helfrich. Working together, nanoENGINEER-1 allows users to interactively define, calculate and visualize 2D electrostatic potential (ESP) images of molecular models. To see an example of this new capability, check out the ESP Image jig in the nanoENGINEER-1 Gallery.
Mac OS X
Alpha 7 does not yet work on Intel-based Mac systems. We are working on this and hope to support Intel-based Mac systems in the coming weeks. We also just discovered a problem plotting nanoENGINEER-1 simulation trace files using GNUplot on Mac OS X. We will be fixing this problem in the next day or two and posting a new MacOS install package in the Download Area.
Saturday, April 15, 2006
Creepy crawlies
When Eric Drexler published Engines of Creation, he included a section on run-away nanotech replicators. The notion is that if there are replicators running around that can use almost anything as feedstock, they buzz through the biosphere converting everything to copies of themselves, leaving behind an ocean of replicators. This is called the "gray goo scenario".
This idea has been popularized by Bill Joy and Michael Crichton and, along with the toxicity of present-day nanoparticles, is considered by many to be one of the grave risks we face in pursuing nanotechnology. Bill Joy's article in Wired recommends that we (the U.S.? the developed nations? Bill and the mouse in his pocket?) simply refrain from developing the technology.
There's a problem: we might refrain, but others won't. The research is difficult but not impossible. Those who pursue the research would be the people who didn't agree to refrain from development, and this dangerous new technology would end up in the hands of those we'd be most fearful of having it.
Drexler and the Foresight Institute, seeking to educate the public and help reason win out over panic, have struggled with these worries for years now. He has argued that building a free-ranging eat-anything replicator is a very difficult engineering problem, comparable to building a car that can forage in the woods for fuel when it runs out of gasoline. Nobody designs a foraging car by accident. A gray-goo replicator may not be possible at all, and if it is possible, it will be the design work of many years.
A dangerous replicator might evolve from nanomachines that started out safe. Many troublesome viruses probably started out as mutations of innocent snippets of DNA. Human-designed nanomachines should not be permitted to evolve. There has been keen interest in nanofactories in recent years, where the instructions are clearly separate from the assembly machinery (what Ralph Merkle calls a broadcast architecture, and computer folks call a SIMD architecture). The instructions are only put to use when the human user decides to push the button to make stuff happen. Autonomous self-replication can not occur, and therefore evolution cannot occur.
Another way to prevent evolution is to ensure that every mutation is fatal. Suppose the replicator's only copy of its blueprint is encrypted, and replication requires decrypting the blueprint each time. If you change one bit or one character in an encrypted document and try to decrypt it, the whole thing becomes meaningless garbage. Any change to the encrypted blueprint will turn the decryption into garbage, and the machine won't be able to build anything.
Finally, there are different ways to pull the plug. One is to stop supplying the energy required for replication. Another is to require tha replication depend upon some exotic "vitamin" available only in a controlled environment.
This idea has been popularized by Bill Joy and Michael Crichton and, along with the toxicity of present-day nanoparticles, is considered by many to be one of the grave risks we face in pursuing nanotechnology. Bill Joy's article in Wired recommends that we (the U.S.? the developed nations? Bill and the mouse in his pocket?) simply refrain from developing the technology.
There's a problem: we might refrain, but others won't. The research is difficult but not impossible. Those who pursue the research would be the people who didn't agree to refrain from development, and this dangerous new technology would end up in the hands of those we'd be most fearful of having it.
Drexler and the Foresight Institute, seeking to educate the public and help reason win out over panic, have struggled with these worries for years now. He has argued that building a free-ranging eat-anything replicator is a very difficult engineering problem, comparable to building a car that can forage in the woods for fuel when it runs out of gasoline. Nobody designs a foraging car by accident. A gray-goo replicator may not be possible at all, and if it is possible, it will be the design work of many years.
A dangerous replicator might evolve from nanomachines that started out safe. Many troublesome viruses probably started out as mutations of innocent snippets of DNA. Human-designed nanomachines should not be permitted to evolve. There has been keen interest in nanofactories in recent years, where the instructions are clearly separate from the assembly machinery (what Ralph Merkle calls a broadcast architecture, and computer folks call a SIMD architecture). The instructions are only put to use when the human user decides to push the button to make stuff happen. Autonomous self-replication can not occur, and therefore evolution cannot occur.
Another way to prevent evolution is to ensure that every mutation is fatal. Suppose the replicator's only copy of its blueprint is encrypted, and replication requires decrypting the blueprint each time. If you change one bit or one character in an encrypted document and try to decrypt it, the whole thing becomes meaningless garbage. Any change to the encrypted blueprint will turn the decryption into garbage, and the machine won't be able to build anything.
Finally, there are different ways to pull the plug. One is to stop supplying the energy required for replication. Another is to require tha replication depend upon some exotic "vitamin" available only in a controlled environment.
Monday, April 10, 2006
Scanning probe microscopy
SPM is a collection of microscopy techniques (of which the first was the scanning tunneling microscope or STM) that permit the imaging of individual atoms. The resolution of the microscopes is not limited by diffraction, but only by the size of the probe-sample interaction volume, which can be as small as a few picometres. The interaction can be used to modify the sample to create small structures.
The STM is an ingenious gadget, invented in 1981 at IBM Zurich. An atomically sharp probe is positioned above a sample; the gap is typically on the order of a nanometer. The probe and the sample are both electrically conductive. A voltage is applied between the probe and the sample. Electrons tunnel across the gap.
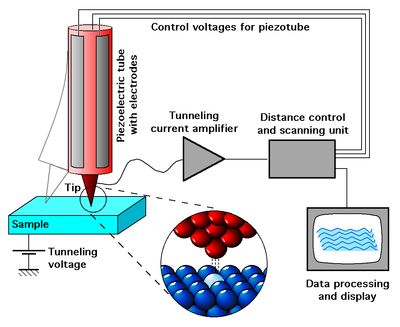
It turns out that the tunneling current is very strongly dependent upon the gap distance. So you build a servomechanism that moves the probe up and down in the Z direction, to keep the gap distance constant by keeping the tunneling current constant. Next, you scan the probe in the X and Y directions, tracking out a raster pattern like the one on your television screen. By looking at the Z position information from the servomechanism, you end up with a function Z = f(X, Y) and when you plot that function, or use it to color the pixels on a computer screen, you get pretty pictures of atoms.

The STM is so conceptually simple that it's possible to build one as a hobbyist.
A few years after the STM, the AFM or atomic force microscope was invented. With this device, the interaction between the probe (this time on the end of a cantilever) and sample is not an electric current flow. Instead it is a force due to the Van der Waals interaction. The force can be measured using piezoelectric strain gages built into the cantilever, or using a reflected laser beam to measure deflection of the cantilever.

You would have hoped that these microscopes would have been useful in pushing atoms around as well as seeing them. To a limited extent they are, but not so useful that you can start to build molecular machines with them. Darn.
The STM is an ingenious gadget, invented in 1981 at IBM Zurich. An atomically sharp probe is positioned above a sample; the gap is typically on the order of a nanometer. The probe and the sample are both electrically conductive. A voltage is applied between the probe and the sample. Electrons tunnel across the gap.

It turns out that the tunneling current is very strongly dependent upon the gap distance. So you build a servomechanism that moves the probe up and down in the Z direction, to keep the gap distance constant by keeping the tunneling current constant. Next, you scan the probe in the X and Y directions, tracking out a raster pattern like the one on your television screen. By looking at the Z position information from the servomechanism, you end up with a function Z = f(X, Y) and when you plot that function, or use it to color the pixels on a computer screen, you get pretty pictures of atoms.

The STM is so conceptually simple that it's possible to build one as a hobbyist.
A few years after the STM, the AFM or atomic force microscope was invented. With this device, the interaction between the probe (this time on the end of a cantilever) and sample is not an electric current flow. Instead it is a force due to the Van der Waals interaction. The force can be measured using piezoelectric strain gages built into the cantilever, or using a reflected laser beam to measure deflection of the cantilever.

You would have hoped that these microscopes would have been useful in pushing atoms around as well as seeing them. To a limited extent they are, but not so useful that you can start to build molecular machines with them. Darn.
Molecular electronics
Over the past few years there has been a lot of fascinating work in molecular electronics. Because of the success of digital logic implemented in VLSI over the past several decades, molecular electronics researchers have focused most of their attention on implementing digital circuits using logic gates.
Early work included the identification of a molecular rectifier or diode, a device that allows electrical current to pass one way but not the other. Arranging diodes in a crossbar arrangement is a major step toward the implementation of complex logic circuits. Additional pieces required to make it practical include nanoscale wires, inverters, and amplifiers, and maybe a flip-flop.

Carbon nanotubes can have different chiralities, which give different electrical properties. Some chiralities give conduction so that the nanotube functions as a wire. Other chiralities give semiconductor behavior, introducing the possibility that a nanotube may be useful as a carbon transistor.
There was some very interesting work done by HP and UCLA in January 2002, basically a way to build a rectangular array of north-south and east-west wires, and connect them by fuses that could be selectively blown. That permits a lot of flexibility in wiring up circuits, which you need to do anything interesting. I haven't heard much more about the HP-UCLA work since 2002, though, and I wonder if they've hit some kind of stumbling block. Their work was based on rotaxanes and catenanes.
There is more to say on this topic, and it will merit another posting one of these days.
Early work included the identification of a molecular rectifier or diode, a device that allows electrical current to pass one way but not the other. Arranging diodes in a crossbar arrangement is a major step toward the implementation of complex logic circuits. Additional pieces required to make it practical include nanoscale wires, inverters, and amplifiers, and maybe a flip-flop.

Carbon nanotubes can have different chiralities, which give different electrical properties. Some chiralities give conduction so that the nanotube functions as a wire. Other chiralities give semiconductor behavior, introducing the possibility that a nanotube may be useful as a carbon transistor.
There was some very interesting work done by HP and UCLA in January 2002, basically a way to build a rectangular array of north-south and east-west wires, and connect them by fuses that could be selectively blown. That permits a lot of flexibility in wiring up circuits, which you need to do anything interesting. I haven't heard much more about the HP-UCLA work since 2002, though, and I wonder if they've hit some kind of stumbling block. Their work was based on rotaxanes and catenanes.
There is more to say on this topic, and it will merit another posting one of these days.
Thursday, April 06, 2006
More molecular machines in nature
Painless blog posting: plug "molecular machine" into Google Scholar, and cut-and-paste whatever comes out! I'm not quite that lazy, I looked at them to see if they were reasonably complete, and weren't of such narrow interest that only three people in the world would want to read them.
Two molecular machines combined
The first combination of two molecular machines has been claimed by Kazushi Kinbara at the University of Tokyo, Japan. The first piece is a light-driven actuator which twists a double bond one way in UV light, and the other way in visible light, causing a pliers-like pair of pieces to hinge closed and open. The ends of the pliers connect to a couple of flippers. By pulsing the light correctly, one can presumably get the flippers to kick, and make the thing swim around in a liquid.

Click for larger image
According to the article, research labs have produced a profusion of interesting molecular machines, but none by itself does anything complex enough to be really useful or extensible. The innovation here is supposed to be the combination of multiple machines. I'm not sure that flapping flippers is much more useful behavior than any of the individual machines would perform. It would be interesting Kinbara had found some kind of general approach by which different submachines could be built into large assemblies, but there is no evidence for that in the articles I could find. Kinbara evidently sees the value of that as a goal, and guesses that large assemblies are about five years out.

Click for larger image
According to the article, research labs have produced a profusion of interesting molecular machines, but none by itself does anything complex enough to be really useful or extensible. The innovation here is supposed to be the combination of multiple machines. I'm not sure that flapping flippers is much more useful behavior than any of the individual machines would perform. It would be interesting Kinbara had found some kind of general approach by which different submachines could be built into large assemblies, but there is no evidence for that in the articles I could find. Kinbara evidently sees the value of that as a goal, and guesses that large assemblies are about five years out.
Sunday, March 26, 2006
More steps along the way
If one thinks about this stuff much, one must inevitably ask, what is the pathway to get from here to there? Ordinarily when attacking a big complicated task, one partitions it into many small subtasks. Then you can draw a big diagram with little boxes connected by lines, the boxes representing subtasks and the lines representing dependencies between subtasks, what the project management weenies call a PERT chart. So where is the PERT chart for developing advanced molecular manufacturing?

Rob Freitas took a stab at one a couple years ago, aimed specifically at diamondoid systems. At the most recent Foresight conference, Drexler and Damian Allis presented work on a tooltip similar in appearance to those described by Freitas, and also intended for extracting and depositing individual atoms on a work surface. More or less simultaneously the Foresight Institute announced their plan to develop a technology roadmap to get us to "productive nanosystems", which is basically the nanofactory shown in the animation. From there it's a relatively small step to any other form of nanotechnology. The nanofactory is the preferred concept today because there is no conceivable way that it could get loose or mutate or go out of control, and people worry about such things.
The people doing Foresight's technology roadmap stuff are talking to people with money, so that might turn out to be interesting. There is also the work by Schafmeister and Rothemund. So it's a pretty interesting time.

Rob Freitas took a stab at one a couple years ago, aimed specifically at diamondoid systems. At the most recent Foresight conference, Drexler and Damian Allis presented work on a tooltip similar in appearance to those described by Freitas, and also intended for extracting and depositing individual atoms on a work surface. More or less simultaneously the Foresight Institute announced their plan to develop a technology roadmap to get us to "productive nanosystems", which is basically the nanofactory shown in the animation. From there it's a relatively small step to any other form of nanotechnology. The nanofactory is the preferred concept today because there is no conceivable way that it could get loose or mutate or go out of control, and people worry about such things.
The people doing Foresight's technology roadmap stuff are talking to people with money, so that might turn out to be interesting. There is also the work by Schafmeister and Rothemund. So it's a pretty interesting time.
Monday, March 20, 2006
DNA origami
There is an MSNBC story about "DNA origami", a technique invented by Paul Rothemund at Caltech to form large complicated shapes by controlling folding patterns in long DNA chains.

Each of the two smiley faces are giant DNA complexes, imaged with an atomic force microscope. Each is about 100 nanometers across (1/1000th the width of a human hair), 2 nanometers thick, and comprised of about 14,000 DNA bases. 7000 of these DNA bases belong to a long single strand. The other 7000 of these bases belong to about 250 shorter strands, each about 30 bases long. These short strands fold the long strand into the smiley face shape. - Paul Rothemund
Rothemund's work is published in Nature, and the full text is available as a PDF at his website. Rothemund works in the DNA and Natural Algorithms Group headed by Erik Winfree.

Each of the two smiley faces are giant DNA complexes, imaged with an atomic force microscope. Each is about 100 nanometers across (1/1000th the width of a human hair), 2 nanometers thick, and comprised of about 14,000 DNA bases. 7000 of these DNA bases belong to a long single strand. The other 7000 of these bases belong to about 250 shorter strands, each about 30 bases long. These short strands fold the long strand into the smiley face shape. - Paul Rothemund
Rothemund's work is published in Nature, and the full text is available as a PDF at his website. Rothemund works in the DNA and Natural Algorithms Group headed by Erik Winfree.
Monday, March 06, 2006
DNA computing progress
On February 24th, National Geographic reported on progress in DNA computing. This field began in 1994 with Leonard Adelman's work on solving the Hamiltonian path problem, and then I didn't hear much about it after that.
Ehud Shapiro and his colleagues at the Weizmann Institute in Israel have developed a DNA computer that can perform 3.3x1014 operations per second. A cubic centimeter of "computer soup" contains about 1.5x1016 individual computers, with a memory capacity of about 6x1020 bytes (ten billion 60-gigabyte hard drives). The two primary advances are that the system is generally programmable, and that the computation is powered by DNA rather than ATP.
"Autonomous bio-molecular computers may be able to work as 'doctors in a cell,' operating inside living cells and sensing anomalies in the host," said Shapiro. "Consulting their programmed medical knowledge, the computers could respond to anomalies by synthesizing and releasing drugs."This approach appears applicable only to embarrassingly parallel problems involving no inter-processor communication. It should be able to tackle some problems in global optimization, though probably not very complicated ones like de-novo protein design, one of the possible pathways toward advanced nanotechnology. For the time being, it's not yet clear that this is a very useful technique.
Monday, January 30, 2006
Design tools for nanotechnology
I've thought a lot about nanotechnology design systems over the past ten years. First I read a beautiful description of a virtual reality system for studying molecular machines up close. It was feasible but not economical; some day that will change. Next I read a little about molecular mechanics, and realized that the principles involved were well within my mathematical and programming abilities.
So I wrote some code (first C, then Lisp, finally Java) that allowed you to put together a little structure with carbon, hydrogen, oxygen, and nitrogen. You could energy-minimize it, and theoretically I could have done equations of motion, but at some point I got busy with other things.
On mailing lists and Usenet, I continued to think more about design systems for nanotech.
So I wrote some code (first C, then Lisp, finally Java) that allowed you to put together a little structure with carbon, hydrogen, oxygen, and nitrogen. You could energy-minimize it, and theoretically I could have done equations of motion, but at some point I got busy with other things.
On mailing lists and Usenet, I continued to think more about design systems for nanotech.
- In the electronics world there is a language for design and simulation called VHDL, which offers a lot of great facilities for managing the complexity of very large projects. Some day nanotech will have its own version of VHDL. Developing it will require learning which details can be safely ignored to give a really terse description of a useful structure.
- When simulating, you don't want to model every single atom in a huge structure. Frequently you want to say that these million atoms just act like a big block, with a little rubberiness or sponginess, but I don't care about their individual vibrations inside the block.
- It would be extraordinarily cool if I could put on VR goggles and force-feedback gloves and physically interact with the structures I design.
- If we accept the premise that real nanotech hardware may be potentially dangerous, we should develop design and simulation software first, so that we're familiar with the dangerous things before they actually exist.
Subscribe to:
Comments (Atom)



